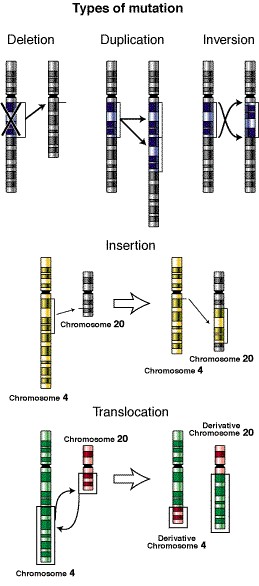
WHAT ARE THE GENOME’S MOBILE ELEMENTS?
Moving elements are pieces of DNA with the ability to jump from one part of the genome to another. While most DNA always stays in the same place, surrounded by the same neighboring pieces of DNA, these moving elements move from one place to another, causing many changes or mutations. We could say that they are like the rebels of the genome.
Some of these changes make organisms more likely to survive and/or reproduce and, therefore, more likely to transmit this change to their offspring.
A mobile element (or “transposable element” or “TE“) is a fragment of DNA that can change its position within the genome as if it were a grasshopper, either directly cutting and sticking itself in another place (transposons) or indirectly by action of an intermediate RNA (retrotransposons). They are present in all living organisms that have been studied, with very few exceptions (such as the bacterium Bacillus subtilis or the parasitic protozoans Leishmania and Plasmodium falciparum). They are more abundant in eukaryotes* than in prokaryotes*. For example, in humans more than two-thirds of the genome are mobile elements; in Drosophila melanogaster, 10-15%, and in corn, 80-85%. In eukaryotes, they can jump from one chromosome to another or jump from one place to another on the same chromosome. In contrast, in prokaryotes, they jump within chromosomal DNA, between a chromosome and a plasmid*, or between two plasmids.
- * Eukaryote: Organisms whose cells have nuclei.
- * Plasmids: Prokaryotic circular DNA molecule.
- * Prokaryote: Organisms formed by a cell that does not have a membrane surrounding the genetic material (that is, without a nucleus).
HOW CAN WE CLASSIFY THEM?

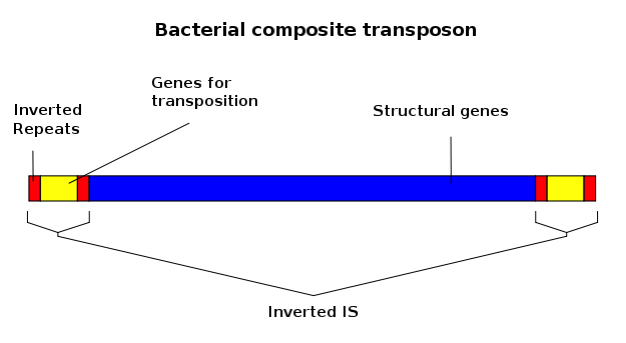
The mobile elements can be classified into two large groups: class I (retrotransposons) and class II (transposons).
Transposons are those elements that move directly from one place to another in the genome using an enzyme (transposase) to cut and paste themselves elsewhere. Two examples are ‘activators’ – transposons that operate independently, and ‘dissociators’ – transposons that depend on triggers. Both were studied by Barbara McClintock* in strains of corn.
On the other hand, there are retrotransposons. As in the case of transposons, the host cell recognizes the retrotransposon as a normal DNA sequence and makes an RNA copy. In the case of retrotransposons, this RNA is converted into DNA using an enzyme called retrotranscriptase. Retrotransposons are sub-classified into two types: viral and non-viral. Two examples of viral retrotransposons are Ty transposons and copy transposons. Non-viral retrotransposons include examples such as LINEs, which encode reverse transcriptase and are transcribed by RNA polymerase II; SINEs, which do not encode reverse transcriptase and are transcribed by RNA polymerase III, and the Alu elements.

 * Barbara McClintock (1902 – 1992) was an American scientist who specialized in cytogenetics, who was awarded the Nobel Prize for Medicine or Physiology in 1983 for her studies and discoveries in corn in the 1940s and 1950s. McClintock discovered the transposition of mobile elements in the genome and used it to explain how genes determine certain physical characteristics. She developed the hypotheses on the regulation of gene expression and the transmission of characters from “mother” plants to “daughter” plants of corn. In addition, due to the high volume of her scientific work, she was awarded many prizes, and she became a member of the National Academy of Sciences of the United States.
* Barbara McClintock (1902 – 1992) was an American scientist who specialized in cytogenetics, who was awarded the Nobel Prize for Medicine or Physiology in 1983 for her studies and discoveries in corn in the 1940s and 1950s. McClintock discovered the transposition of mobile elements in the genome and used it to explain how genes determine certain physical characteristics. She developed the hypotheses on the regulation of gene expression and the transmission of characters from “mother” plants to “daughter” plants of corn. In addition, due to the high volume of her scientific work, she was awarded many prizes, and she became a member of the National Academy of Sciences of the United States.
HOW CAN THEY AFFECT THE GENOME?
Transposable elements can cause different changes or mutations within the genome.
For starters, when a transposon moves through the genome it can insert into a vital gene, disrupting its function; or into a location near vital genes, causing erroneous activation or inactivation.
On the other hand, chromosomal rearrangements can occur when transposons of the same family, with complementary sequences that are physically separated in the genome, match up due to sequence homology. This match-up can lead to ectopic recombination*. Large-scale rearrangements induced by mobile elements are considered a particular class of recombination events that influence genomic plasticity; they can cause inversions, duplications or even deletions of up to one hundred kilobases.
One of the greatest contributions of transposable elements to the evolution of the host genome is to provide a source of genetic material to create new genes and new functions. Transposable elements have numerous properties that predispose them to be integrated into the host genome, providing functional advantages.
- * Ectopic Recombination: Recombination between sequences in non-sister chromatids (i.e. chromosome arm).
WHAT EFFECTS CAN THEY CAUSE?
These modifications in the genome can have either positive, negative or neutral effects.
There are a large number of mutations that can be harmful and cause diseases, such as hemophilia A and B, neurofibromatosis, or familial hypercholesterolemia, which are caused by insertions of Alu elements or by ectopic recombination of different copies of SINEs.
 On the other hand, an example of a beneficial mutation happens in telomeres*, consisting of tandem repeats of two retrotransposons HeT-A and TART. Its transpositional activity compensates for the absence of the enzyme telomerase. This can be seen in Drosophila melanogaster.
On the other hand, an example of a beneficial mutation happens in telomeres*, consisting of tandem repeats of two retrotransposons HeT-A and TART. Its transpositional activity compensates for the absence of the enzyme telomerase. This can be seen in Drosophila melanogaster.
An example of a neutral mutation can be seen in petunias, where the genetic line W138 originates from the wild line R27. The main difference between the two is that W138 contains a mutation in the AN1 locus, responsible for the production of flower pigments, which results in an irregular coloration.
- * Telomere: end of the chromosome.
HOW IS IT POSSIBLE THAT WITH SO MANY MOBILE ELEMENTS, THAT CAN CAUSE SO MANY MUTATIONS, WE ARE STILL ALIVE?
Different mechanisms regulate and control mobile elements. These mechanisms are common to some organisms and specific to others. However, they are always present.
There are general control mechanisms in eukaryotic genomes which result in gene silencing that can act at different levels. First, transcriptional silencing acts on the promoter regions of genes, blocking transcription. Post-transcriptional silencing works at the RNA level, degrading or blocking its translation.
 An example of gene silencing that we find in Drosophila melanogaster, among others, is Piwi-associated RNAs (piRNAs). The piRNAs are generated from regions of the genome where there are different inserted mobile elements. When this region is transcribed, a transcript is formed containing different copies of the moving elements. This RNA is cut into small pieces, of 24-31 nucleotides, called piRNAs. These piRNAs bind to a Piwi group protein, which leads the piRNA to the complementary mRNA (messenger RNA) sequence of the mobile element of the same family. A Piwi-associated enzyme breaks down TE mRNAs and releases a piRNA chain, which can associate with other Piwi proteins. The binding of this complex in the initial transcript containing different copies of the mobile elements regenerates a piRNA complementary to the previous one that can be directed to the TE mRNA. This is known as a “ping pong cycle.”
An example of gene silencing that we find in Drosophila melanogaster, among others, is Piwi-associated RNAs (piRNAs). The piRNAs are generated from regions of the genome where there are different inserted mobile elements. When this region is transcribed, a transcript is formed containing different copies of the moving elements. This RNA is cut into small pieces, of 24-31 nucleotides, called piRNAs. These piRNAs bind to a Piwi group protein, which leads the piRNA to the complementary mRNA (messenger RNA) sequence of the mobile element of the same family. A Piwi-associated enzyme breaks down TE mRNAs and releases a piRNA chain, which can associate with other Piwi proteins. The binding of this complex in the initial transcript containing different copies of the mobile elements regenerates a piRNA complementary to the previous one that can be directed to the TE mRNA. This is known as a “ping pong cycle.”
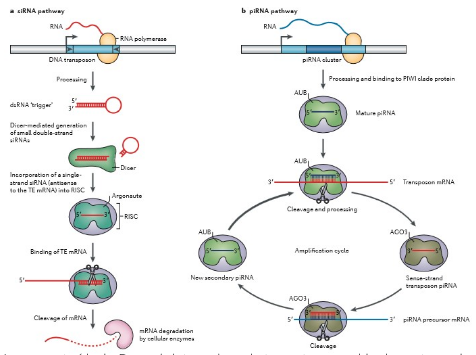
On the other hand, there are also other mechanisms present in humans such as siRNA (small interfering RNA) or interference RNA (RNAi). These, like piRNAs, function by sequence homology. First, the transcript is formed as for piRNAs. This transcript has the peculiarity to be composed of a sequence with inverted long terminations, which are complementary to each other and can match together, forming a double-stranded hairpin-shaped RNA structure. The cell does not recognize the double RNA as its own, so by means of a protein (Dicer), it eliminates the loop. Once this is done, one of the two chains is degraded and the other chain binds to another protein (Argonaut). At this point, the complex called RISC is formed. The chain is paired by sequence homology to an mRNA, which is subsequently degraded by an enzyme.
Interference RNA can be initiated by exposing cells to RNA either through transfection* or endogenous (i.e. internal) expression. In mammalian systems, the effect of sequence-specific RNAi has been observed by expression of 21-23 base transcripts capable of duplexing, or by expression of short hairpin RNA.
Another mechanism of gene silencing is DNA methylation, which involves the addition of a methyl group to cytosine nucleotides in DNA. There are two hypotheses about how methylation can affect gene transcription. First, DNA methylation itself can physically impede the binding of proteins necessary for gene transcription, standing between them and DNA. To transcribe DNA, the chromatin must be decompacted so that the genetic information can be read. The second hypothesis states that proteins are added to the methylated DNA, compacting the chromatin and giving rise to heterochromatin. This binding results in a structural change that impedes access to the gene that needs to be transcribed.
In conclusion, the control of transposition is essential for the survival of organisms. However, this control does not imply the definitive inactivation of the effects of certain moving elements.
- * Transfection: introduction of external genetic material into cells using plasmids or viral vectors.
Writing: Henry Estrella and Mar Gutiérrez, students of the “Youth and Science” Program who made their stay at González Lab